Downloading..
New! The spider is split into a core package and into additional extra algorithm modules which can be downloaded seperately from here.
The core package of spider can be downloaded here .Just
unzip into the directory of choice. Start MATLAB and run the script
"use_spider" to startup. Additional algorithms have to be extracted
into the same directory.
If you use the Spider all the time, it would be a good idea to call
"use_spider" in your startup.m file and to add the Spider root
directory permanentlyto your MATLAB search path using, for example,
PATHTOOL. A provided program "spider_init" attempts to do this but if
your startup.m file is not in the usual place (in the matlab directory)
then you haveto do this manually.
Some objects require WEKA to be installed. Go to the WEKA page and download the current release of WEKA. You have to put the weka.jar file into the directory
matlabroot \ java \ jar
Note that WEKA is a java library, thus you have to use the JVM of matlab
running. This is by default the case EXCEPT you launch it with the flag -nojvm.
In this case, WEKA wont work.
If you call "use_spider", you should get a message
> WEKA support enabled!
Once installed, to get started type "help spider" and "help svm" or try out the tutorial.
or use toy
and
toy2d
, simple objects for generating toy datasets:
Simple examples to start with...
Want to try something simple to start with on the command-line? Try these:
X=rand(50)-0.5; Y=sign(sum(X,2)); r=train(svm,data(X,Y))
loss(train(svm,toy))
loss(train(cv(svm),toy))
get_mean(train(cv(svm),toy))
k=param(knn,'k',1:5); get_mean(train(cv(group({k gridsel(k) svm})),toy))
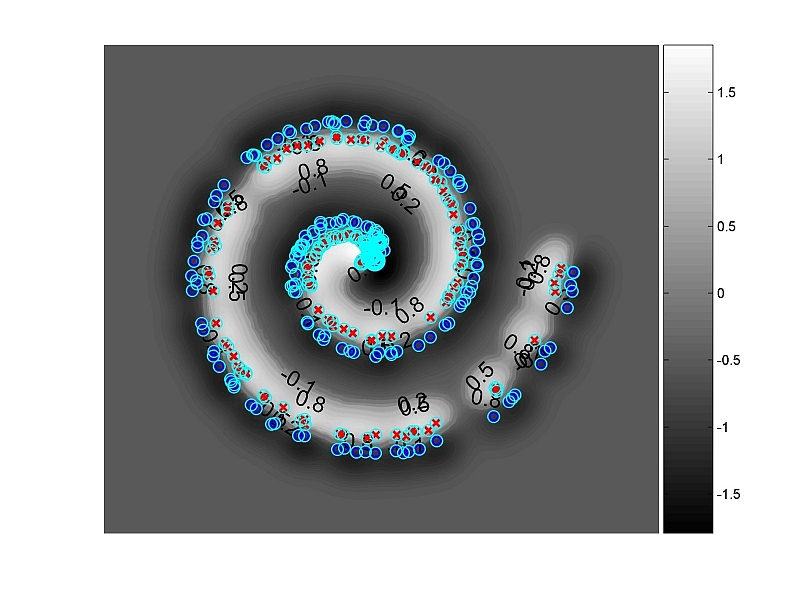
or use spiral to generate the famous toy-benchmark:
d=gen(spiral({'m=200','n=2','noise=0.35'}));
[r,a]=train(svm({kernel('rbf',1),'optimizer="andre"'}),d)
plot(a)
and get: